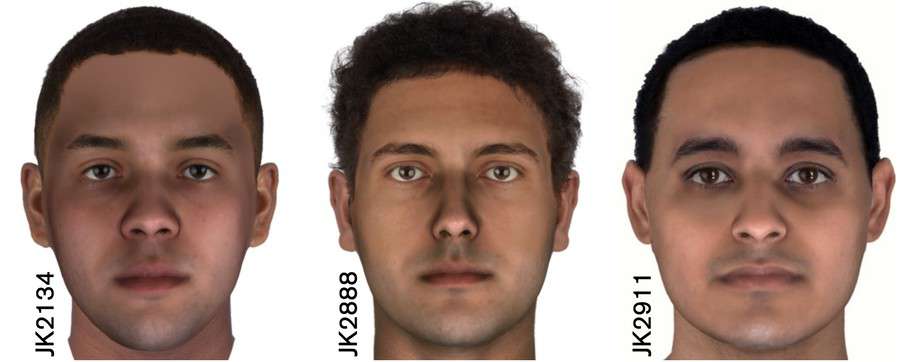
At the 32nd International Symposium on Human Identification (ISHI), being held this week in Orlando, FL, Parabon NanoLabs will unveil for the first time the predicted faces of three ancient mummies from an ancient Nile community in Egypt known as the Abusir el-Meleq. The mummy samples, estimated to be between 2,023 and 2,797 years old, were processed by researchers from the Max Planck Institute for the Science of Human History and the University of Tubingen in Germany (Schuenemann et al. 2017).1 Enzymatic damage repair was performed on each sample, after which they were sequenced with a capture assay targeting 1.24 million single nucleotide polymorphisms (SNPs) and aligned to the human reference genome. Parabon used the resulting whole-genome sequencing data, which is publicly available in the European Nucleotide Archive (ENA), and selected three samples with the highest quality data to analyze. The company believes this is the first time comprehensive DNA phenotyping has been performed on human DNA of this age, and the results are being presented to the forensic community in ISHI poster #51.Learn More About Snapshot DNA Analysis »
This work was made possible by recent bioinformatic advances in the field of low-coverage imputation, which allows for highly accurate determination of common SNP genotypes from low-coverage sequencing data. The sequencing coverages for the three mummy samples were only 0.13X, 0.23X, and 0.96X, respectively, meaning that each position in the genome was covered less than one time on average. Because individuals have two copies of each SNP, only 0.1%, 0.3%, and 2.8% of the SNPs, respectively, could be directly called from the data, which would not be enough information for phenotype prediction or for traditional imputation. With low-coverage imputation, however, the SNP call rates jumped to 29%, 39%, and 65%, respectively. Parabon bioinformaticist and WGS analyst, Dr. Janet Cady, who spearheaded the work, is enthusiastic about the results. “Parabon has been the leader in forensic microarray analysis for years, and with the introduction of this new imputation technology, we can now handle even the most challenging samples, ancient or forensic” she said.See All Published Police Investigations »
After the imputation step, Parabon applied its Snapshot DNA Phenotyping pipeline to each of the three mummy samples. Snapshot was designed to operate on challenging forensic samples, so it is specifically calibrated to handle missing data, some of which remains even after low-coverage imputation. Snapshot predicted each mummy’s ancestry, pigmentation, and face morphology. Interestingly, their ancestry was determined to be more similar to modern Mediterranean and Middle Eastern individuals than to modern Egyptians. Their complexions were predicted to be light brown, with dark eyes and hair and no freckles. These results are highly consistent with Schuenemann et al’s conclusions that “ancient Egyptians shared more ancestry with Near Easterners than present-day Egyptians, who received additional sub-Saharan admixture in more recent times” and that they had an allele for lighter skin. Three-dimensional face morphology was inferred by predicting the values of face principal components (PCs), which were then transformed into 3D graphical meshes. The face predictions were compared to one another, and heat maps were calculated to show the differences between the subjects. These differences were then emphasized to create caricatured faces, which were combined with the pigmentation predictions to create composites of the individuals’ likely appearance at age 25 by Parabon’s forensic artist.Read More Snapshot Success Stories »
“It’s great to see how genome sequencing and advanced bioinformatics can be applied to ancient DNA samples,” said Dr. Ellen Greytak, Parabon’s Director of Bioinformatics. “Just like in Parabon’s law enforcement casework, these techniques are revolutionizing ancient DNA analysis because they operate on fragmented DNA and have been shown to be sensitive down to only 10 picograms of DNA.”
A print-ready PDF of the scientific poster can be downloaded from Parabon’s website at https://snapshot.parabon.com/ishi2021-poster .
.
from ScienceBlog.com https://ift.tt/39Qq3UE
No comments:
Post a Comment