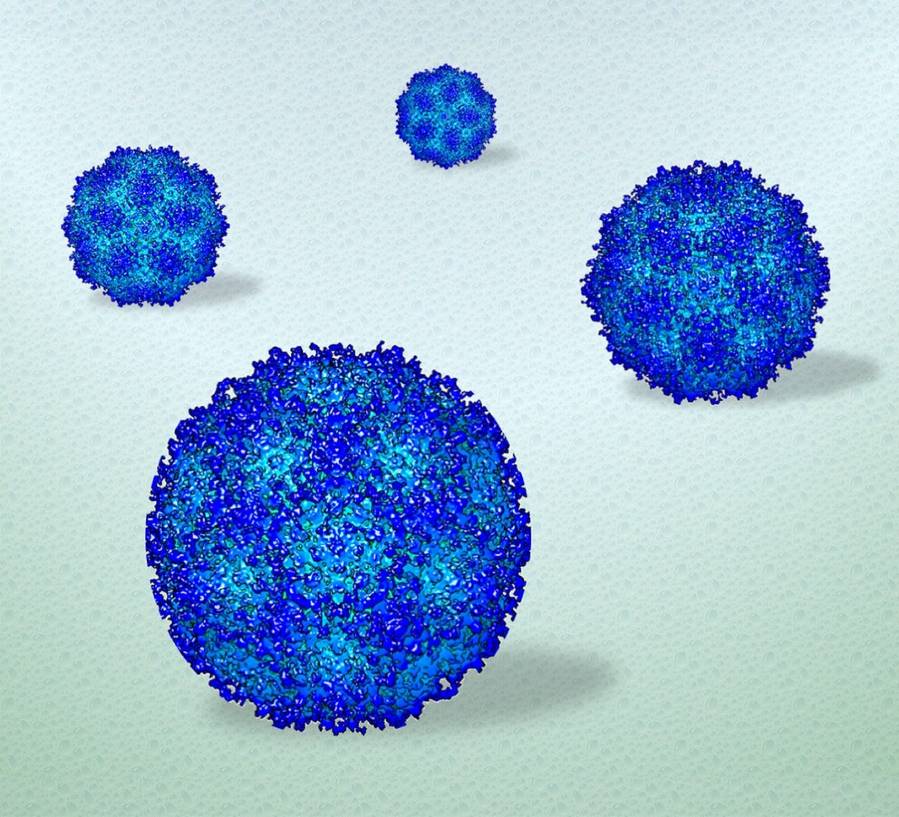
A pond in summer can reveal more about a fish than a pond in winter. The fish living in icy conditions might remain still enough to study its scales, but to understand how the fish swims and behaves, it needs to freely move in three dimensions. The same holds true for analyzing how biological items, such as viruses, move in the human body, according to a research team led by Deb Kelly, Huck Chair in Molecular Biophysics and professor of biomedical engineering at Penn State, who has used advanced electron microscopy (EM) technology to see how human viruses move in high resolution in a near-native environment. The visualization technique could lead to improved understanding of how vaccine candidates and treatments behave and function as they interact with target cells, Kelly said.
In an effort to expand the tools scientists have to study the microscopic world, researchers recorded live, 20-second-long movies of human viruses floating in liquid at near-atomic detail in an electron microscope. The same degree of information, immediately available as they record, may take up to 24 hours to acquire using traditional static imaging methods. Their approach and results were made available online July 24 in Advanced Materials.
“The challenge remained to view biological materials in dynamic systems that reflects their authentic performance in the body,” said Kelly, who also directs the Penn State Center for Structural Oncology. “Our results show new structures and active insights of human viruses contained in minute volumes of liquid — the same size as respiratory droplets that spread SARS-CoV-2.”
Cryogenic electron microscopy (cryo-EM) is becoming the field’s gold standard for observing samples at or beyond atomic resolution, according to Kelly. The technique involves flash freezing the sample and focusing a beam of electrons through it. The electrons and the sample’s components interact, which is captured by detectors embedded in the instrument. Thousands of images can be processed to calculate what the item looks like in 3D — but more is needed to fully understand how the item functions in a more natural setting.
“While cryo-EM can tell us a lot of information, it still produces a static image,” said GM Jonaid, the paper’s first author and a student in the Bioinformatics and Genomics Graduate Program in the Huck Institutes of the Life Sciences. Jonaid is conducting his doctoral dissertation research in Kelly’s lab. “With improved chips and a powerful direct detector on the microscope, we can accumulate a lot of movie frames to view how the sample acts in real time. We can see things how they exist — not just how we prepared them.”
The researchers used adeno-associated virus (AAV) as a model system to demonstrate their approach. AAV is a biological nanoparticle that can be used to help deliver vaccines or treatments directly to cells. The platform is based on a hijacked adenovirus, which can easily enter several kinds of cells. The ease with which it interacts with cells makes it a useful capsule to transport its engineered payload.
“AAV is a well-known, gene therapy vehicle with current applications involved in drug delivery and vaccine development for COVID-19,” Kelly said. “This model system is already well-studied so we can use it to validate our approach with the goal of seeing biological entitles in a liquid state, as maintained in the human body.”
The researchers applied minute volumes of liquid solution containing AAV to the wells of specialized silicon nitride microchips, commercially supplied by Protochips Inc. They then placed the microchip assemblies in the EM to examine the viruses in action.
“The images are very comparable to cryo-EM data, but the preparation was less complex, less technically involved,” Jonaid said. “Once we had the images, taken rapidly, like frames of a movie, we processed them just like we would any other high-resolution data.”
The results were videos of AAV moving in liquid, with subtle changes in the particle’s surface, suggesting that the particle’s physical properties change as it explores its environment, Kelly said. The resolution was close to three to four Angstroms (a single atom is measured as one Angstrom).
Once they proved the imaging strategies worked, the researchers set their sights on a smaller target: antibodies produced by COVID-19 patients.
“We saw how antibodies contained in the serum of COVID-19 patients interacted with the remaining SARS-CoV-2 particles,” Kelly said, noting that the ability to observe such interactions would be especially useful when assessing the viability of vaccine candidates prior to clinical trials.
Kelly and her team plan to continue investigating the molecular underpinnings of SARS-CoV-2 and host-receptor proteins using liquid phase-EM, as a complement to the information garnered from cryo-EM results.
“You really need data from both techniques to understand how viruses look and behave in the living body,” Kelly said. “Visualizing the dynamic movement in solution complements high-resolution snapshots to reveal more complete information.”
Other contributors include William Dearnaley and Michael Casasanta, researchers in Kelly’s lab; Liam Kaylor, student in the Penn State Molecular, Cellular, and Integrative Biosciences Graduate Program and research assistant in Kelly’s lab; Samantha Berry, graduate student, Penn State Department of Biomedical Engineering; Jennifer Gray, staff scientist and assistant research professor, Penn State Materials Research Institute; Madeline Dukes, Protochips Inc.; and Michael Spilman, Direct Electron LP.
The National Institutes of Health and the National Cancer Institute supported this work.
from ScienceBlog.com https://ift.tt/2WrfbsT
No comments:
Post a Comment